The Kabat Numbering Scheme
The Kabat numbering scheme is a widely adopted standard for numbering the residues in an antibody in a consistent manner. However the scheme has problems!
First, since the numbering scheme was developed from sequence data (a fairly limited set), the position at which insertions occur in CDR-L1 and CDR-H1 does not match the structural insertion position. Thus topologically equivalent residues in these loops do not get the same number.
Second, the numbering adopts a rigid specification. For example in the potentially very long CDR-H3, insertions are numbered between residue H100 and H101 with letters up to K (i.e. H100, H100A ... H100K, H101). If there are more residues than that, there is no standard way of numbering them. Such situations occur at other positions too.
The numbering throughout the chains is as follows:
Light chain
0 1 2 3 4 5 6 7 8 9
10 11 12 13 14 15 16 17 18 19
20 21 22 23 24 25 26 27
27A 27B 27C 27D 27E 27F 28 29
30 31 32 33 34 35 36 37 38 39
40 41 42 43 44 45 46 47 48 49
50 51 52 53 54 55 56 57 58 59
60 61 62 63 64 65 66 67 68 69
70 71 72 73 74 75 76 77 78 79
80 81 82 83 84 85 86 87 88 89
90 91 92 93 94 95
95A 95B 95C 95D 95E 95F 96 97 98 99
100 101 102 103 104 105 106
106A 107 108 109
Heavy chain
0 1 2 3 4 5 6 7 8 9
10 11 12 13 14 15 16 17 18 19
20 21 22 23 24 25 26 27 28 29
30 31 32 33 34 35
35A 35B 36 37 38 39
40 41 42 43 44 45 46 47 48 49
50 51 52
52A 52B 52C 53 54 55 56 57 58 59
60 61 62 63 64 65 66 67 68 69
70 71 72 73 74 75 76 77 78 79
80 81 82
82A 82B 82C 83 84 85 86 87 88 89
90 91 92 93 94 95 96 97 98 99
100
100A 100B 100C 100D 100E 100F 100G 100H 100I 100J
100K 101 102 103 104 105 106 107 108 109
110 111 112 113
The Chothia Numbering Scheme
The Chothia numbering scheme is identical to the Kabat scheme, but places the insertions in CDR-L1 and CDR-H1 at the structurally correct positions. This means that topologically equivalent residues in these loops do get the same label (unlike the Kabat scheme).
There are two disadvantages: first, the Kabat scheme is so widely used that some confusion can arise; second, Chothia et al. changed their numbering scheme as of their 1989 Nature paper such that insertions in CDR-L1 are placed after residue L31 rather than L30. Examining the conformations of the loops shows that L30 is the correct position.
Note That in their latest paper (Al-Lazikani et al., (1997) JMB 273,927-948), Chothia's group returns to using residue L30 as the insertion site in CDR-L1!
The pre-1989/post-1997 Chothia numbering (the structurally correct version) throughout the chains follows.
Light chain
0 1 2 3 4 5 6 7 8 9
10 11 12 13 14 15 16 17 18 19
20 21 22 23 24 25 26 27 28 29
30
30A 30B 30C 30D 30E 30F
31 32 33 34 35 36 37 38 39
40 41 42 43 44 45 46 47 48 49
50 51 52 53 54 55 56 57 58 59
60 61 62 63 64 65 66 67 68 69
70 71 72 73 74 75 76 77 78 79
80 81 82 83 84 85 86 87 88 89
90 91 92 93 94 95
95A 95B 95C 95D 95E 95F 96 97 98 99
100 101 102 103 104 105 106
106A 107 108 109
Heavy chain
0 1 2 3 4 5 6 7 8 9
10 11 12 13 14 15 16 17 18 19
20 21 22 23 24 25 26 27 28 29
30 31
31A 31B
32 33 34 35 36 37 38 39
40 41 42 43 44 45 46 47 48 49
50 51 52
52A 52B 52C 53 54 55 56 57 58 59
60 61 62 63 64 65 66 67 68 69
70 71 72 73 74 75 76 77 78 79
80 81 82
82A 82B 82C 83 84 85 86 87 88 89
90 91 92 93 94 95 96 97 98 99
100
100A 100B 100C 100D 100E 100F 100G 100H 100I 100J
100K 101 102 103 104 105 106 107 108 109
110 111 112 113
Martin (Enhanced Chothia) Numbering Scheme
The only differences between the Chothia and Kabat numbering schemes are in the sites of indels in CDR-L1 and CDR-H1.
Our 'Martin' (Enhanced Chothia) scheme also considers the structurally correct locations for indels in the framework regions. Thus the numbering scheme is identical to the Chothia in most regards but positions of framework indels have been refined.
The most important of these is the insertion which present in the majority of antibodies at H82a,b,c has been moved to the structurally correct location of H72a,b,c.
We have also introduced an indel site at L52 in CDR-L2. All structures have the standard length of 7 residues and the conformations are relatively conserved. However sequences of varying length are seen and analysis of the structure suggests this is the correct location. It also corresponds with the AHo numbering scheme.
In this scheme, the locations of deleted residues are indicated with ().
A manuscript describing this new scheme is in preparation.
Light chain
0 1 2 3 4 5 6 7 8 9
(10) 11 12 13 14 15 16 17 18 19
20 21 22 23 24 25 26 27 28 29
(30)
30A 30B 30C 30D 30E 30F
31 32 33 34 35 36 37 38 39
40
40A (41) 42 43 44 45 46 47 48 49
50 51 (52)
52A 52B 52C 52D 52E
53 54 55 56 57 58 59
60 61 62 63 64 65 66 67 (68)
68A 68B 68C 68D 68E 68F 68G 68H 69
70 71 72 73 74 75 76 77 78 79
80 81 82 83 84 85 86 87 88 89
90 91 92 93 94 (95)
95A 95B 95C 95D 95E 95F 96 97 98 99
100 101 102 103 104 105 106 107
107A 108 109
110
Heavy chain
0 1 2 3 4 5 6 7 (8)
8A 8B 8C 8D 9
10 11 12 13 14 15 16 17 18 19
20 21 22 23 24 25 26 27 28 29
30 (31)
31A 31B
32 33 34 35 36 37 38 39
40 41 (42) 43 44 45 46 47 48 49
50 51 (52)
52A 52B 52C 53 54 55 56 57 58 59
60 61 62 63 64 65 66 67 68 69
70 71 72
72A 72B 72C 73 74 75 76 77 78 79
80 81 82 83 84 85 86 87 88 89
90 91 92 93 94 95 96 97 98 99
(100)
100A 100B 100C 100D 100E 100F 100G 100H 100I 100J
100K 101 102 103 104 105 106 107 108 109
110 111 112 113
Table of CDR Definitions
A number of definitions of the CDRs are commonly in use:
- The Kabat definition is based on sequence variability and is the most commonly used
- The Chothia definition is based on the location of the structural loop regions - see more detail at the bottom of this section
- The AbM definition is a compromise between the two used by Oxford Molecular's AbM antibody modelling software
- The contact definition has been recently introduced by us and is based on an analysis of the available complex crystal structures. This definition is likely to be the most useful for people wishing to perform mutagenesis to modify the afinity of an antibody since these are residues which take part in interactions with antigen. Lists of CDR residues making contact in each antibody with summary data for each CDR
| Loop | Kabat | AbM | Chothia1 | Contact2 | IMGT |
|---|---|---|---|---|---|
| L1 | L24-L34 | L24-L34 | L26-L32 | L30-L36 | L27-L32 |
| L2 | L50-L56 | L50-L56 | L50-L52 | L46-L55 | L50-L51 |
| L3 | L89-L97 | L89-L97 | L91-L96 | L89-L96 | L89-L97 |
| H1 | H31-H35B (Kabat Numbering)3 | H26-H35B | H26-H32..34 | H30-H35B | H26-H35B |
| H1 | H31-H35 (Chothia/Martin Numbering) | H26-H35 | H26-H32 | H30-H35 | H26-H33 |
| H2 | H50-H65 | H50-H58 | H52-H56 | H47-H58 | H51-H56 |
| H3 | H95-H102 | H95-H102 | H96-H101 | H93-H101 | H93-H102 |
Note (1) some of these definitions (particularly for Chothia
loops) vary depending on the individual publication examined
- see below.
28 June 2021: This page previously described the Chothia
CDRs using a wider defintion (taken from these papers), but I have
updated that interpretation to use a consensus. The previous
set was:
CDR-L1:L24-34; CDR-L2:L50-56; CDR-L3:L89-97;
CDR-H1:H26-32; CDR-H2:H52-56; CDR-H3:H95-102.
Note (2) any of the numbering schemes can be used for these CDR defintions, except the contact definition uses the Chothia or Martin (Enhanced Chothia) definition.
Note (3) the end of the Chothia CDR-H1 loop when numbered using the Kabat numbering convention varies between H32 and H34 depending on the length of the loop. (This is because the Kabat numbering scheme places the insertions at H35A and H35B.)
- If neither H35A nor H35B is present, the loop ends at H32
- If only H35A is present, the loop ends at H33
- If both H35A and H35B are present, the loop ends at H34
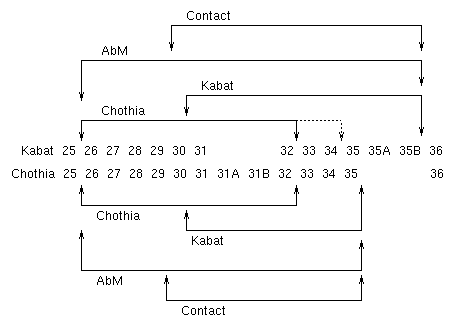
This diagram illustrates the alternative definitions for CDR-H1. The Kabat and Chothia numbering schemes are shown horizontally and the Kabat, Chothia, AbM and Contact definitions of the CDRs are shown with arrows above and below the two numbering schemes.
Chothia CDR definitions
As mentioned above, various papers from Chothia appear to have somewhat different definitions for the CDRS - especially if one considers the figures as well as the text. This table attempts to summarize these.
| Paper | L1 | L2 | L3 | H1 | H2 | H3 | |
|---|---|---|---|---|---|---|---|
| Chothia et al. 1986, Science 233:755-758 | 26-32 | 50-52 | 91-96 | 26-32 | 53-55 | 96-101 | |
| Chothia & Lesk 1987, JMB 196:901-917 | Text | 26-32 | 50-52 | 91-96 | 26-32 | 53-55 | 96-101 |
| Table 6 | 26-33 | 50-52 | 91-96 | 26-32 | 53-55 | 96-101 | |
| Tramontano et al. 1989, PROT 6:823-394 | 26-33* | ? | 91-96 | 26-32 | ? | 95-101+ | |
| Chothia et al. 1989, Nat 342:877-883 | Table 2 | 26-32 | 49-53 | 90-97 | 26-32 | 52a-56 | ? |
| Tramontano et al. 1990, JMB 215:175-182 | ? | ? | ? | ? | 52a-55 | ? | |
| Chothia et al. 1992, JMB 227:799-817 | ? | ? | ? | 26-32 | 52-56 | ? | |
| Barre et al. 1994, NatSB1:915-920 | Table 1 | 25-33 | 50-52 | 90-96 | 26-32 | 52-56 | ? |
| Fig 2 | 25-32 | 50-52 | 91-96 | 26-33 | 52-56 | ? | |
| Tomlinson et al. 1995, EMBO 14:4628-4638 | 26-32 | 50-52 | 91-96 | ? | ? | ? | |
| Al-Lazikani et al. 1997, JMB 273:927-948 | 25-32 | 50-52 | 91-96 | 26-32 | 52-56 | 95-102 | |
| Figs | 25-33 | 49-53 | 90-97 | 25-33 | 50-58 | 92-104 | |
| Morea et al. 1997, BiophyChem 68:9-16 | Fig 2 | ? | ? | ? | ? | ? | 92-104 |
| Morea et al. 1998, JMB 275:269-294 | ? | ? | ? | ? | ? | 92-104 | |
| Consensus interpretation | 26-32 | 50-52 | 91-96 | 26-32 | 52-56^ | 96-101 |
* inferred from length
+ Taken from Figure 10 and text saying that 94 and 102 are the end of the beta-strands
^ Possibly the most controversial: it's very clearly H52a-H55 in some of the papers.
How to identify the CDRs by looking at a sequence
The following set of rules will allow you to find the (Kabat or Chothia) CDRs in an antibody sequence. Note that the word 'always' should always be treated with care! There are rare examples where these virtually constant features do not occur (for example the human heavy chain sequence EU does not have Trp-Gly after CDR-H3). The Cys residues are the best conserved feature.
CDR-L1
- Start
- Approx residue 24
- Residue before
- always a Cys
- Residue after
- always a Trp. Typically Trp-Tyr-Gln, but also, Trp-Leu-Gln, Trp-Phe-Gln, Trp-Tyr-Leu
- Length
- 10 to 17 residues
CDR-L2
- Start
- always 16 residues after the end of L1
- Residues before
- generally Ile-Tyr, but also, Val-Tyr, Ile-Lys, Ile-Phe
- Length
- always 7 residues (except NEW (7FAB) which has a deletion in this region)
CDR-L3
- Start
- always 33 residues after end of L2 (except NEW (7FAB) which has the deletion at the end of CDR-L2)
- Residue before
- always Cys
- Residues after
- always Phe-Gly-XXX-Gly
- Length
- 7 to 11 residues
CDR-H1
- Start
- Approx residue 26 (always 4 after a Cys)
[Chothia / AbM defintion];
Kabat definition starts 5 residues later - Residues before
- always Cys-XXX-XXX-XXX
- Residues after
- always a Trp. Typically Trp-Val, but also, Trp-Ile, Trp-Ala
- Length
- 10 to 12 residues [AbM definition];
Chothia definition excludes the last 4 residues
CDR-H2
- Start
- always 15 residues after the end of Kabat / AbM definition) of CDR-H1
- Residues before
- typically Leu-Glu-Trp-Ile-Gly, but a number of variations
- Residues after
- Lys/Arg-Leu/Ile/Val/Phe/Thr/Ala-Thr/Ser/Ile/Ala
- Length
- Kabat definition 16 to 19 residues;
AbM (and recent Chothia) definition ends 7 residues earlier
CDR-H3
- Start
- always 33 residues (sometimes 30 residues) after the end of CDR-H2 (always 3 after a Cys)
- Residues before
- always Cys-XXX-XXX (typically Cys-Ala-Arg)
- Residues after
- always Trp-Gly-XXX-Gly
- Length
- 3 to 25(!) residues - and sometimes much longer, particularly in bovine antibodies
Table of mean contact data
Following an analysis of the contacts between antibody and antigen in the complex structures available in the Protein Databank, we have generated a set of mean contact data. The full method by which these results were obtained is described in the following paper: MacCallum, R. M., Martin, A. C. R. and Thornton, J. T. 'Antibody-antigen interactions: Contact analysis and binding site topography' J. Mol. Biol. 262:732-745.
Briefly, we analysed the number of contacts made at each position, defining contact as burial by > 1 square Angstrom change in solvent accessibility. These data give a simple measure of how likely a residue is to be involved in antigen contact.
Second, we calculated the mean percentage burial over the accessible residues.
The 'Mean Contacts' table presents the chain name, residue number (pre-1989 Chothia Numbering), the number of contacts and the mean percent burial.
An 'Simplified View' is a list of CDR residues making contact in each antibody with summary data for each CDR.
Mean Contacts Simplified View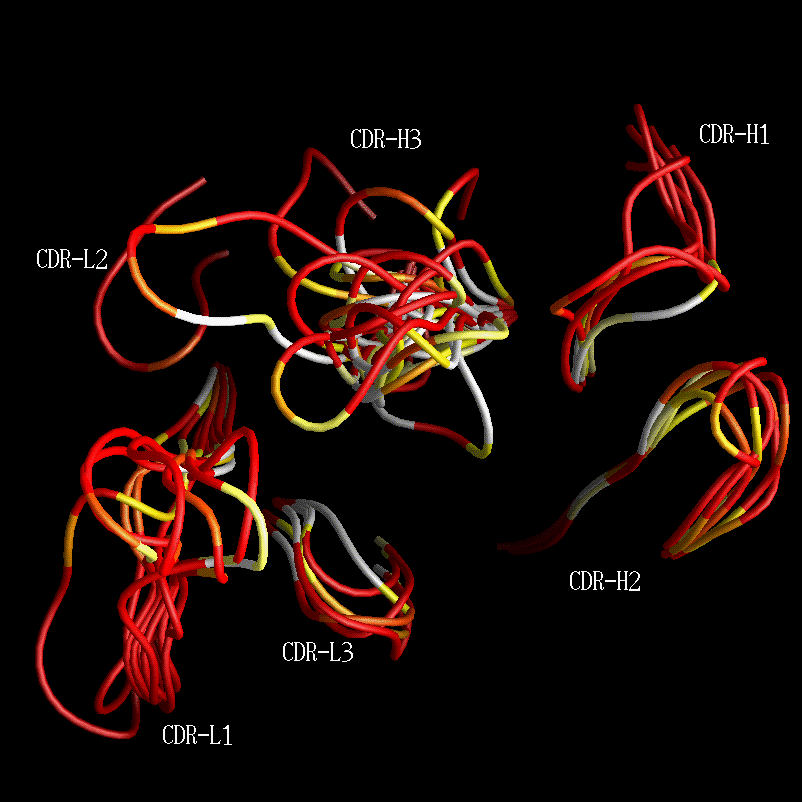
Composite combining site containing all CDR conformations from 1986 coloured by contact propensity.