Peptide Bonds and Protein Backbones
In this part of the practical, you will now download a peptide and view it using RasMol. You will identify and highlight the peptide bonds and measure the torsion angles.
Introduction - The peptide bond
The peptide bond links amino acids. The bond is delocalized:
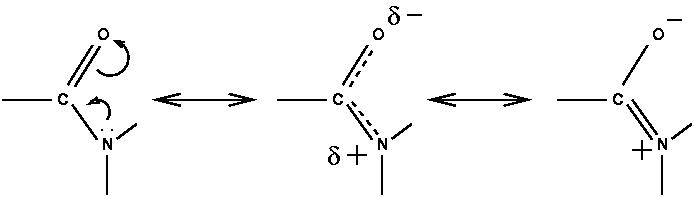
Delocalization means that the peptide bond has partial double-bond character and therefore cannot be rotated freely and is therefore cis (close to 0 degrees) or trans (close to 180 degrees)
Backbone torsion angles
A torsion angle (or 'dihedral' angle) is the 'twist' angle along a bond:
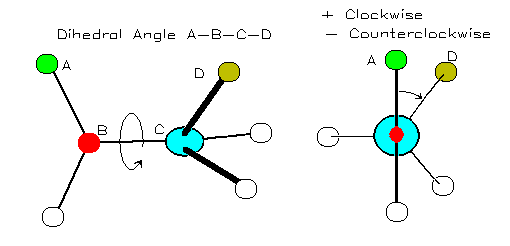
Image from http://bmbiris.bmb.uga.edu/wampler/tutorial/prot2.html
The protein backbone can be described in terms of the phi, psi and omega torsion angles of the bonds:
- The phi angle is the angle around the -N-CA- bond (where 'CA' is the alpha-carbon)
- The psi angle is the angle around the -CA-C- bond
- The omega angle is the angle around the -C-N- bond (i.e. the peptide bond)
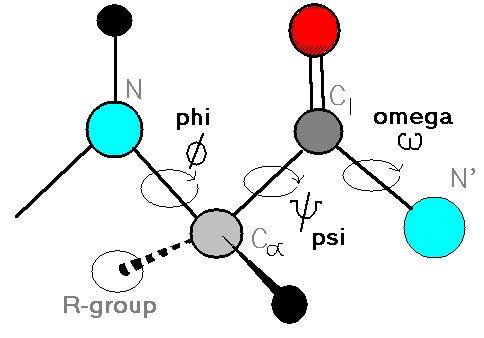
Image from http://bmbiris.bmb.uga.edu/wampler/tutorial/prot2.html
Practical work
First, view this peptide using RasMol. You will see a fragment from the protein crambin with the sequence Thr-Gly-Cys-Ile-Ile.
- Look at the peptide. Identify the backbone nitrogens (coloured in blue) and the backbone carbonyl groups
- Work your way along the backbone and identify each C-alpha atom and its sidechain
- Now type the following commands in RasMol:
select *.c or *.n
wireframe 30
NOTE! This is 2 commands on 2 lines. Type them exactly as you see them - the 'or' does not mean type one or the other, it is part of the command - You will now see each peptide bond highlighted with a thicker line
Your view should look something like the view on the right. If it
doesn't, you have probably messed with the Display
options first! If this is the case, type the command:
select *
The from the menus, click:
Display->Wireframe
Now repeat the stages as described above to get the required view.
Having identified where the peptide bonds are, we will have a look at the torsion angles - since there are no prolines, you would expect these to be trans. Let's see if they are:
- Type the following commands in RasMol:
select *.ca
spacefill 100 - Now rotate the view so that each peptide bond in turn is orientated such that it is coming out of the screen. (i.e. the cylinder of the thick bond is viewed end-on and looks like a small circle).
- You should see that the bonds to the adjacent C-alpha atoms (which are now shown as spheres) are at approximately 180degrees to one another.
Now let's measure the torsion angles to see how close to 180degrees they really are:
- Type the following command in RasMol:
set picking torsion - Find the threonine residue at the N-terminus of the peptide (threonine has an oxygen - in red - in its sidechain and the backbone will have a nitrogen at the end of the chain - in blue)
- Click, in turn:
- the CA of the Thr
- the carbonyl carbon of the Thr
- the nitrogen of the second amino acid (Gly)
- the CA of the Gly
- The torsion angle will be displayed
- Work along the peptide, calculating the omega torsion angle for each of the 4 peptide bonds.
You should have found that all the peptide bonds were indeed in the trans conformation with omega angles close to 180degrees.